Hi there - I am a PhD candidate in Biomedical Informatics at Stanford University, studying artificial intelligence and clinical informatics. I am currently advised by Serena Yeung, Curtis Langlotz, Nigam Shah and previously by Matthew P. Lungren. I am affiliated with Stanford’s MARVL lab and AIMI Center, and have experience working at Google Research , Microsoft Research , Salesforce AI research and The ChanZuckerberg Initiative.
My research focuses on the intersection of multimodal and self-supervised learning, and the application of these methods to improve healthcare.
Featured Publications
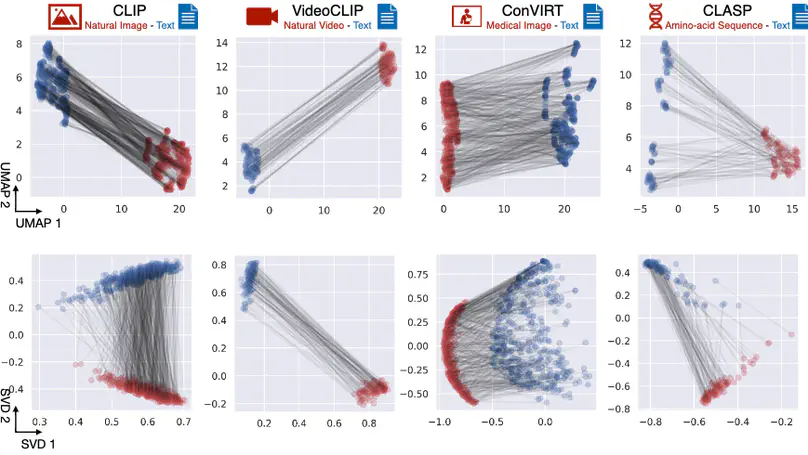
The traditional process of diagnosing model behaviors in deployment settings involves labor-intensive data acquisition and annotation. Our proposed method, DrML, can discover high-error data slices, identify influential attributes and further rectify undesirable model behaviors, without requiring any visual data. Through a combination of theoretical explanation and empirical verification, we present conditions under which classifiers trained on embeddings from one modality can be equivalently applied to embeddings from another modality.
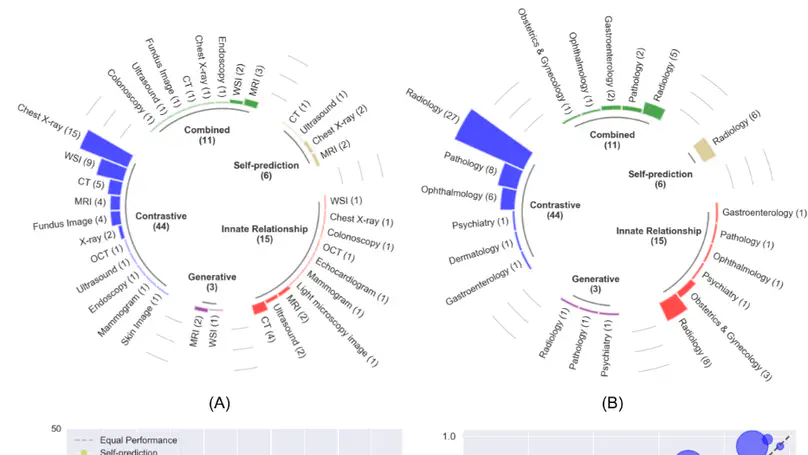
In this review, we provide consistent descriptions of different self-supervised learning strategies and compose a systematic review of papers published between 2012 and 2022 on PubMed, Scopus, and ArXiv that applied self-supervised learning to medical imaging classification. With this comprehensive effort, we synthesize the collective knowledge of prior work and provide implementation guidelines for future researchers interested in applying self-supervised learning to their development of medical imaging classification models.
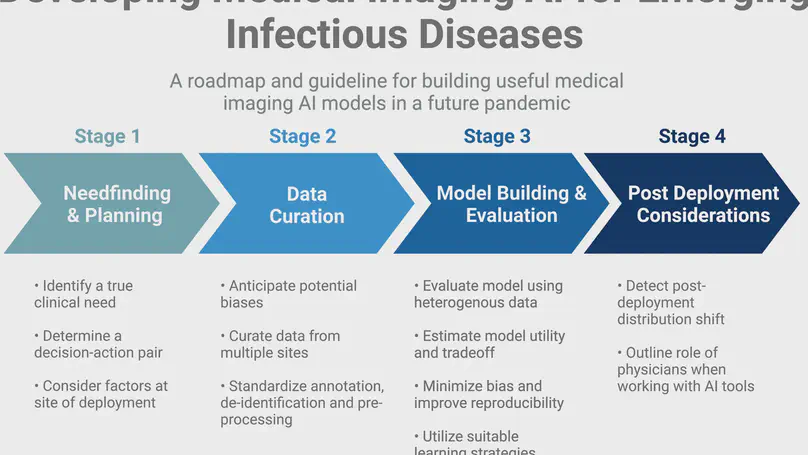
In this review, we provide an evidence-based roadmap for how machine learning technologies in medical imaging can be used to battle ongoing and future pandemics. Specifically, we focus in each section on the four most pressing issues, namely - needfinding, dataset curation, model development and subsequent evaluation, and post-deployment considerations.
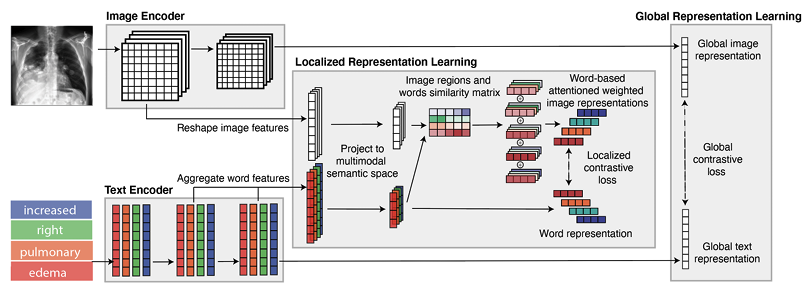
The purpose of this work is to develop label-efficient multimodal medical imaging representations by leveraging radiology reports. Specifically, we propose an attention-based framework (GLoRIA) for learning global and local representations by contrasting image sub-regions and words in the paired report. In addition, we propose methods to leverage the learned representations for various downstream medical image recognition tasks with limited labels.
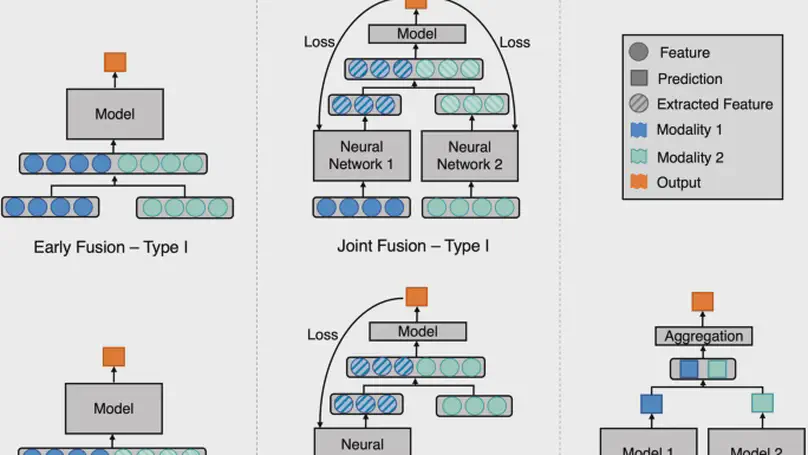
In this reivew, we describe different data fusion techniques that can be applied to combine medical imaging with EHR, and systematically review medical data fusion literature published between 2012 and 2020. By means of this systematic review, we present current knowledge, summarize important results and provide implementation guidelines to serve as a reference for researchers interested in the application of multimodal fusion in medical imaging.
All Publications
Experience
Designed object embeddings to improve dense semantic understanding in Vision Language Models (VLMs) and curated a multi-image Visual Question Answering dataset to benchmark VLM’s dense semantic understanding
Developed a VLM for generating radiology reports from Chest X-rays that achieves state-of-the-art performance
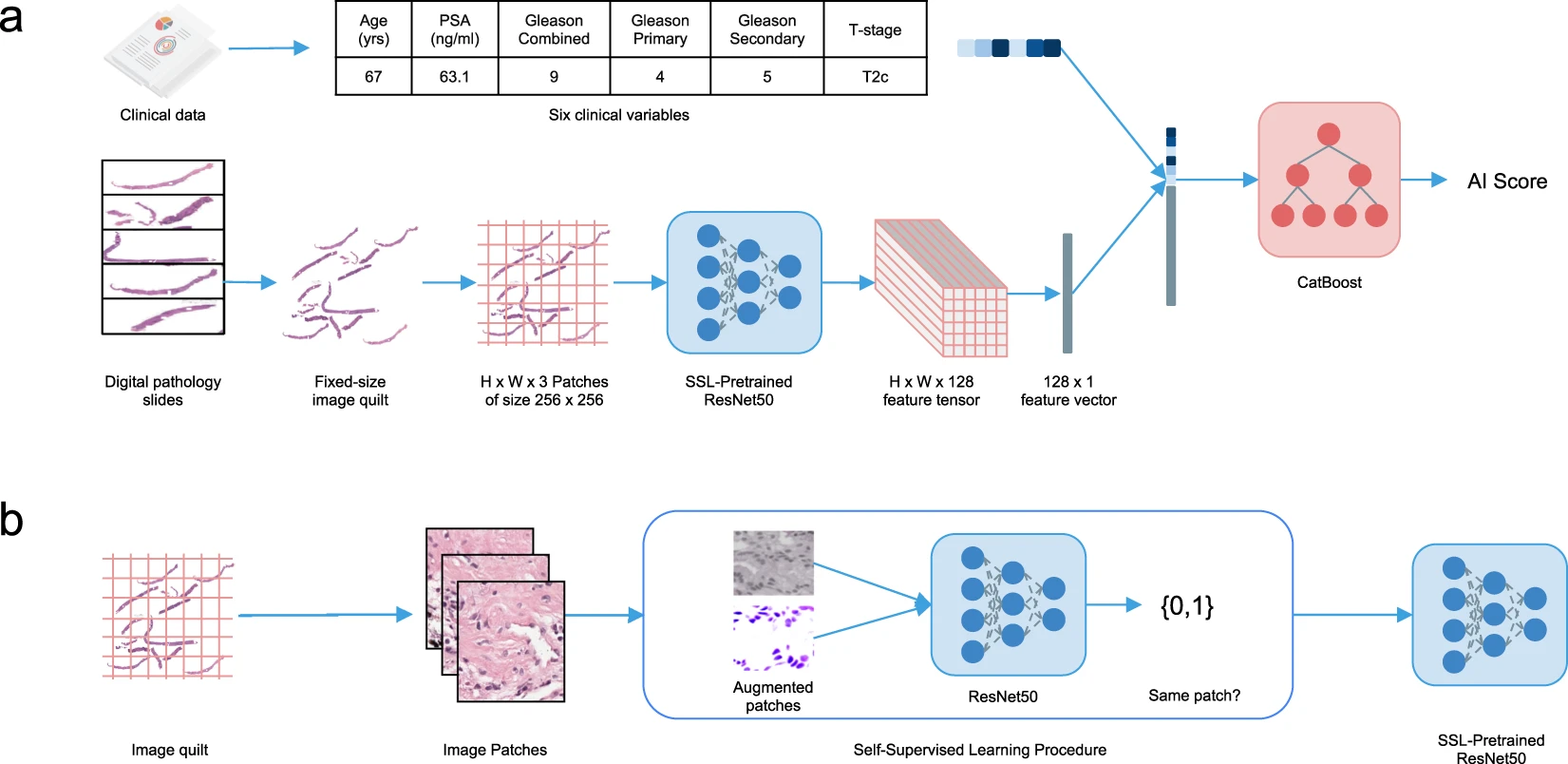
Designed and implemented a multimodal self-supervised framework for prostate cancer long-term outcome prediction.
Created Segmentify, an interactive and general-purpose cell segmentation plugin for the image viewer Napari
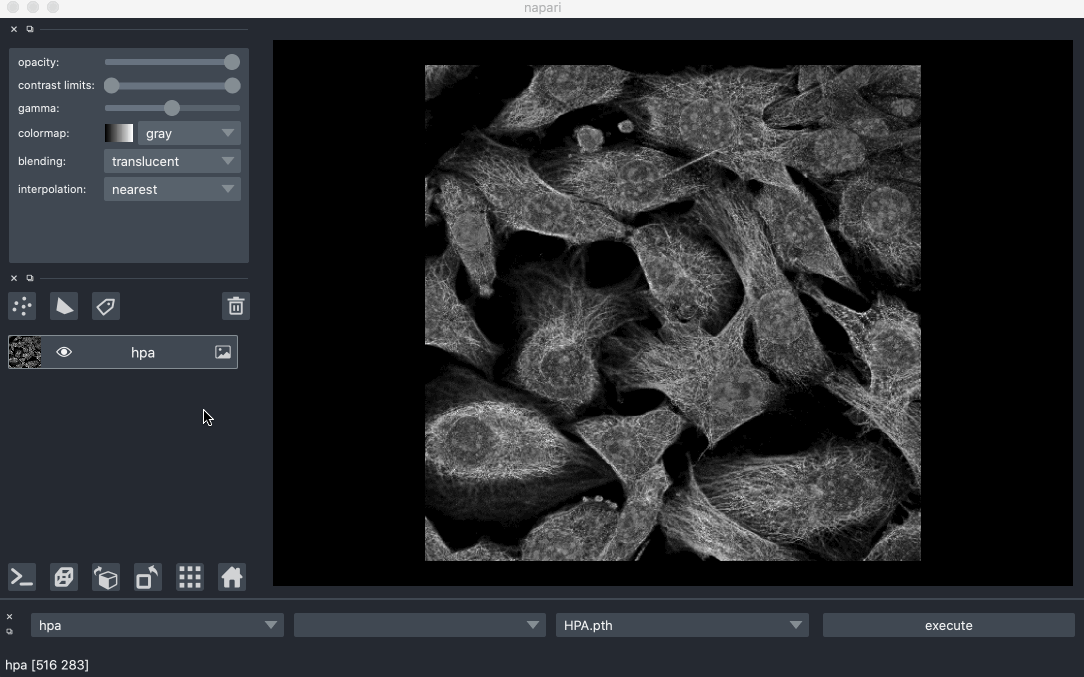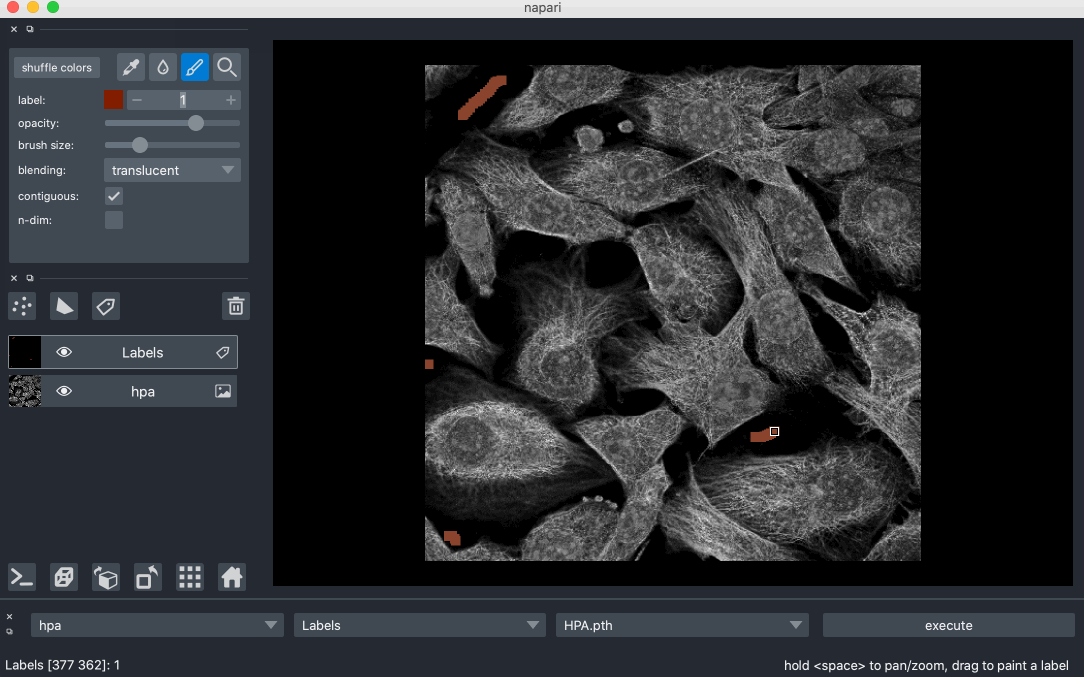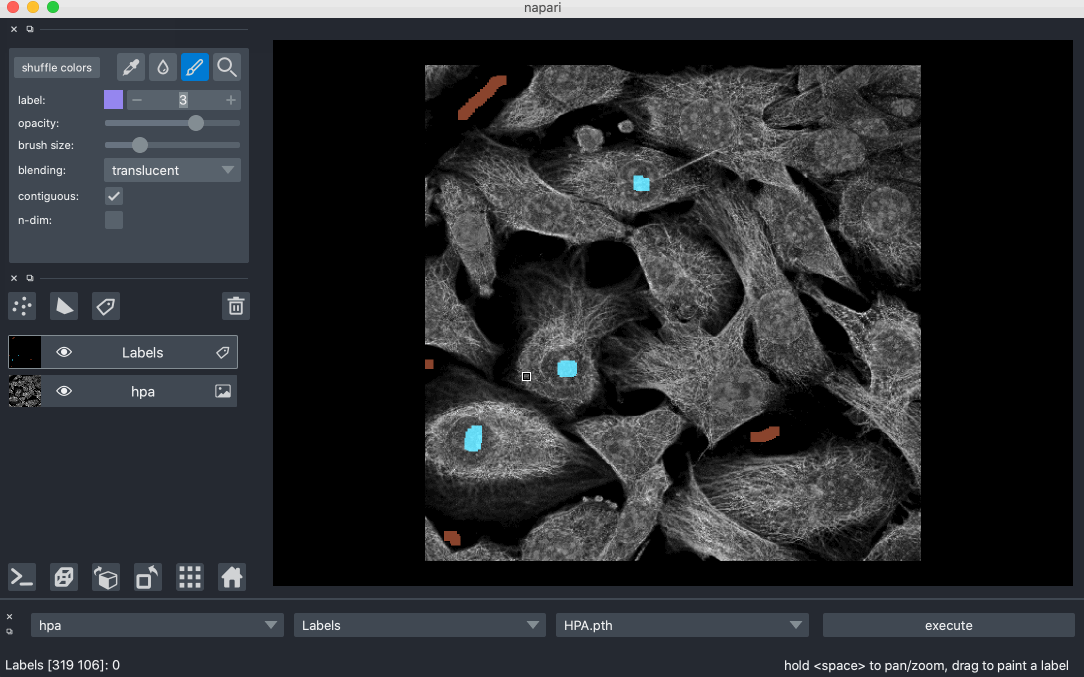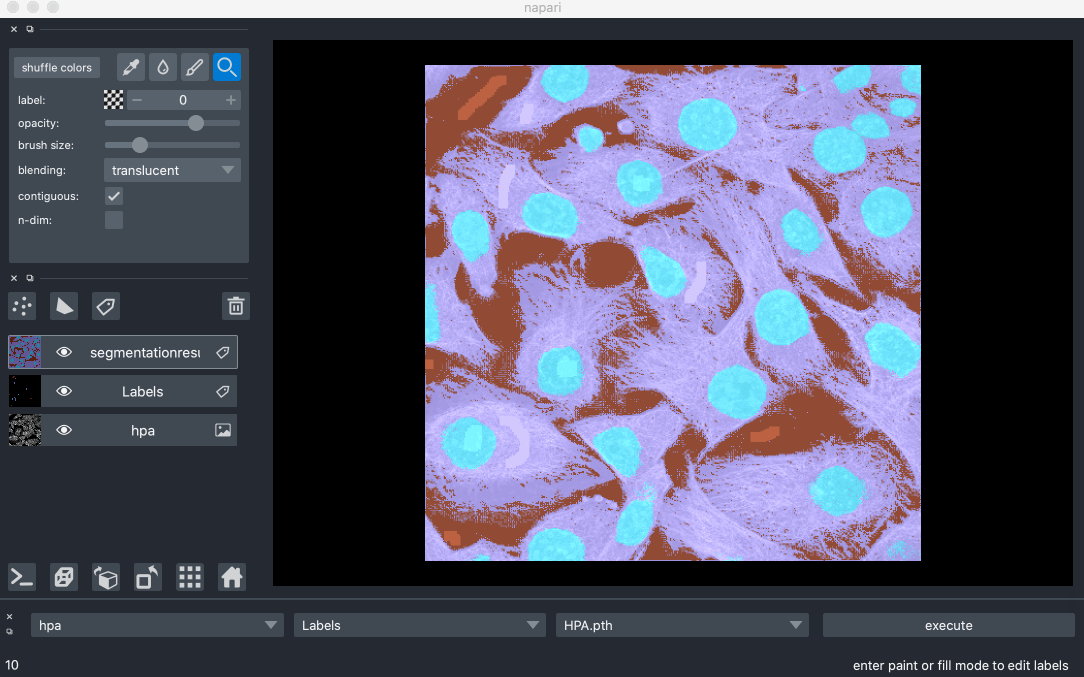
Developed mmtf-pyspark, a python package that parallelizes analysis and mining of protein data using Apache-Spark